How It Works
- Ontological terms have unique, persistent identifiers and logically defined terms, which aids in standardization and clarity.
- The graph-structure (imagine edges and nodes) allow for interence, which aids in discoverability and findability.
- The graph-structure also allows for scalability, including increasing template terms and trait terms.
The purpose of FuTRES is to make individual-level trait data findable, accessible, interoperable, and reuseable. We serve trait data from biological and paleontological specimens in a format that is standardized. The datastore uses an ontological backbone to improve discoverability and promotes novel research. Here is a quick introduction to FuTRES.
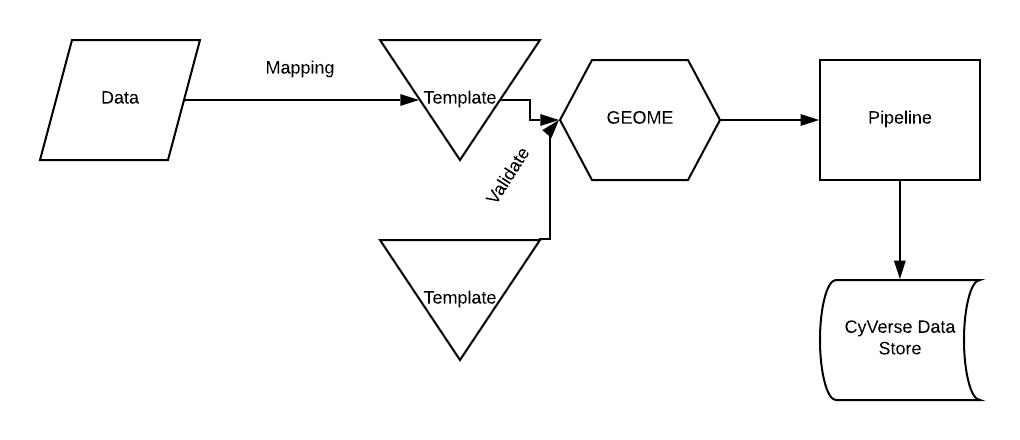
Data Pipeline
The data processing pipeline is comprised of five main steps: pre-processing, triplifying, reasoning, conversion to a tabular format, and data loading. For more information, you can watch this short video on the FOVT Data Pipeline.
The pre-processing step is to help with data standardization. The template (below) helps users format their data. We have also created a RShiny App to help users format their data.
After pre-processing, RDF triples are generated by reading configuration files from each data source that includes term mappings, data validation, and creating relationships between processes and objects as defined by the ontology. All triples are referenced by globally unique identifiers by appending record identifiers from the input data to globally unique, resolvable HTTP prefixes that can be customized for each project. Because instance identifiers are derived from the input data, output identifiers can be linked back to the specific records in the raw source data, which also provides a mechanism to track data provenance.
The next step in the workflow - reasoning - uses the bundled Ontology Development Kit that supports multiple description logic profiles through multiple reasoners. The workflow provides an optional configuration file to OntoPilot that further allows users to customize the reasoning process. The reasoning helps with more flexible searches; for example, searching for "length" will provide all the length terms from the ontology that are in the FuTRES datastore.
The reformatting workflow converts the data to a series of CSV files via a customizable SPARQL query through query_fetcher, a bundled package for fast conversion of RDF to tabular data that is built upon the Apache Jena Java Library. The output data can be loaded into whatever data storage system the user prefers, including key/value stores (e.g., ElasticSearch), relational databases (e.g., PostgreSQL), or triplestores (e.g., Blazegraph).
Pre-processing
Template
We have developed a template (viewable here) to help data providers create datasets that are ready for ingestion into the FuTRES knowledge base. The field names in the template largely correspond to Darwin Core terms. Since Darwin Core is the most commonly used standard for sharing biodiversity occurrence data, these fields may already be pre-existing in most collections databases, or if not, they can be easily mapped or crosswalked from other existing fields.
Formatting data
We have also created a RShiny App to help users format their data in the correct format for uplad to GEOME. You can read more about the RShiny App here and how to contribute data here.
Reasoning
Ontology
An ontology is a knowledge representation which describes concepts and their relationships to one another in a logical framework that is understandable by machines. The benefits of using ontologies are:
The ontology is used for triplifying and reasoning in the data pipeline.
Biological Collections Ontology
The data model relies heavily on the Biological Collections Ontology. The classes in the BCO connects the properties (column headers) in the template to the values in the dataset.
FuTRES Ontology of Vertebrate Traits
The FuTRES Ontology of Vertebrate Traits (FOVT) is an application ontology specifically designed to serve the purposes of the FuTRES projects. It was developed by Dr. Ramona Walls, Dr. Meghan Balk, and Laura Brenskelle, and it reuses many existing ontologies (for example, UBERON, PATO, BSPO, and OBA) to create and define vertebrate traits. These become the controlled vocabulary for "measurementType".
Trait terms can be requested here.
Data Loading
The FuTRES data store serves dataset where each row is a unique measurement (long format). This differs from how researchers may normally collect data, where each row is a specimen. Having each row as a measurement makes it easier for the pipeline as well as for subsequent analyses. Each measurement (row) has a unique diagnosticID. These diagnosticIDs of the same element are connected to each other via materialSampleID. The materialSampleIDs (elements) of the same indiby individualID. As an example, a dataset with four measurements, two from the illium and two from the femur, all belonging to the individual would look like this:
| individualID | materialSampleID | catalogNumber | diagnosticID | scientificName | measurementType | measurementValue | measurementUnit |
|---|---|---|---|---|---|---|---|
| 1 | 1 | FMNH PR2081 | 1 | Tyrannosaurus rex | illium length | 1525 | mm |
| 1 | 1 | FMNH PR2081 | 2 | Tyrannosaurus rex | illium depth | 608 | mm |
| 1 | 2 | FMNH PR2081 | 3 | Tyrannosaurus rex | femur length | 1321 | mm |
| 1 | 2 | FMNH PR2081 | 4 | Tyrannosaurus rex | femur circumference | 580 | mm |
Accessing the datastore
API
The datastore can be accessed through our API, where users can search the datastore and download data.
R Package
The datastore can also be access through the rfutres package (read more here).
